User Manual
1. Quick Start
We provide a quick entrance for users on the HOME page, you can directly
enter the DNA sequence containing the editing site and editing pattern.
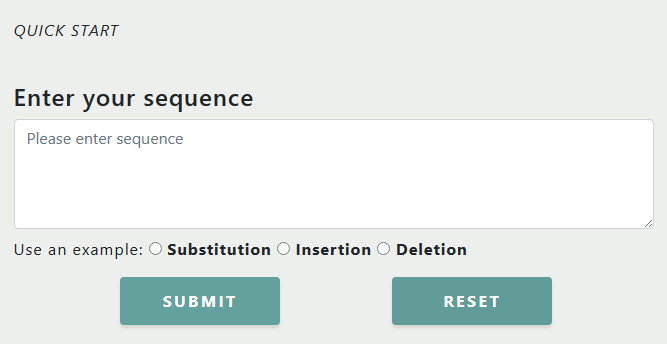
For the input sequence, several rules should be followed:
- Only "ATCGatcg" and "+-/()"are acceptable.
- The bases to be edited are marked with a pair of parentheses "()".
- For Substitutions, the backslash "/" is used. Replacing base X by base Y should be written X/(Y). Only replace-one-base-by-one-base is allowed.
- For Insertions, the plus sign "+" is used. X(+Y) means insert a base Y after base X.
- For Deletions, the minus sign "-" is used. Deleting several bases XYZ after base X should be written X(-XYZ)
For example:
- Example for substitution:
CGCCGCCGCCTCTACTGGGGCTTCTTCTCGGGCCGCGGCCGCGTCAAGCCGGGGGGGCGCTGGCGCGA(/T)GGCCGCCTGGCAACTCTGCGACTACTACCTGCC - Example for insertion:
CGCCGCCGCCTCTACTGGGGCTTCTTCTCGGGCCGCGGCCGCGTCAAGCCGGGGGGGCGCTG(+ATT)GCGCGAGGCCGCCTGGCAACTCTGCGACTACTACCTGCC - Example for deletion:
CGCCGCCGCCTCTACTGGGGCTTCTTCTCGGGCCGCGGCCGCGTCAAGCCGGGGGGGC(-GCTGGCGCGA)GGCCGCCTGGCAACTCTGCGACTACTACCTGCC
We provide users with quick examples. After clicking the radio button of substitution, insertion, or deletion, the corresponding example sequence will be autofilled the text input box.
2. Full Functions
OPED is a webserver tool that supports three design methods for Optimizing Prime Editing Designing, including Design by Sequence, Design by Position, and Search in OPEDVar.
2.1 Design by Sequence
The user can access the Design by Sequence Page through two ways:
a) Navigation Bar -> DESIGN -> BY SEQUENCE;
b) HOME Page > "Input Sequence" Button.
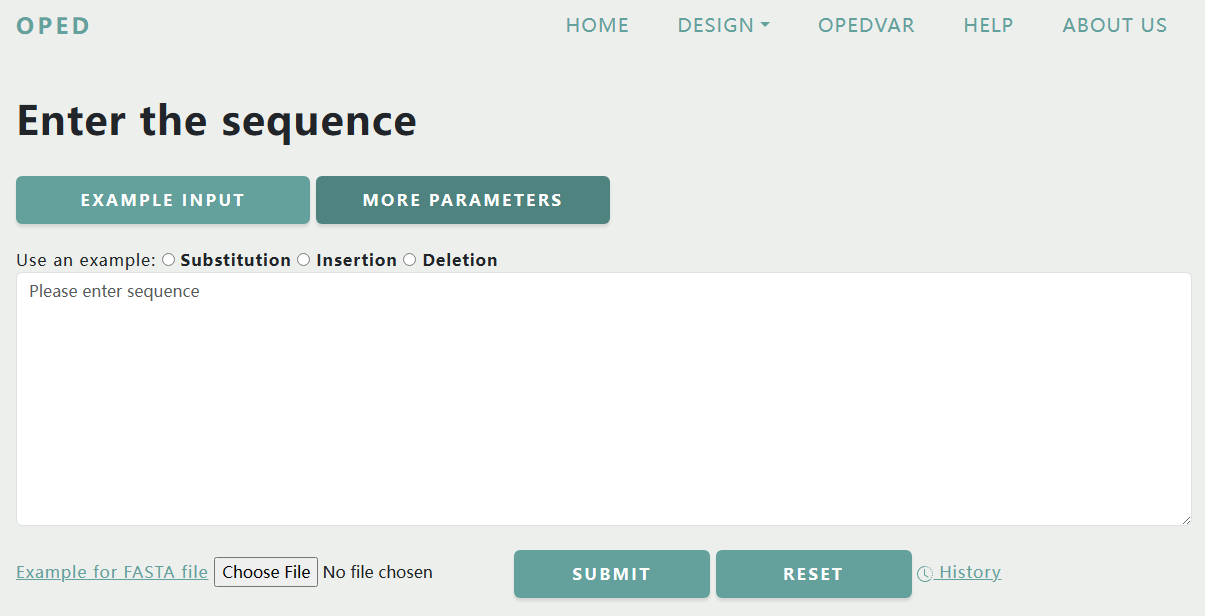
Here, users can predict pegRNAs and sgRNAs by inputting their sequences and setting customized parameters. Users can enter the DNA sequence directly, or submit a FASTA format file. You can view the rules for the input sequence by clicking button "Example Input", and check the parameters by "More Parameters".
In this project, 11 parameters are involved in, including PAM, Maximum Edit-to-Nick Distance, Maximum PBS Length, Minimum PBS Length, Maximum RTT Length, Minimum RTT Length, Maximum Nick-to-Nick Distance, Minimum Nick-to-Nick Distance, Minimum Downstream Homology, Number of Optimized pegRNAs, and Assembly . The initial default parameters are already given and the user can adjust them as needed.

We provide users with quick examples. After clicking the radio button of substitution, insertion, or deletion, the corresponding example sequence will be autofilled into the text input box.
For FASTA files, we follow the general formatting requirements of FASTA files, while requiring the sequence content to meet our rules which are described above. We provide an example FASTA file that can be downloaded by clicking on the "Example for FASTA file" link.
2.2 Design by Position
Users can access the Design by Position Page through two ways:
a) Navigation Bar -> DESIGN -> BY POSITION;
b) HOME Page -> "Input Position" Button.
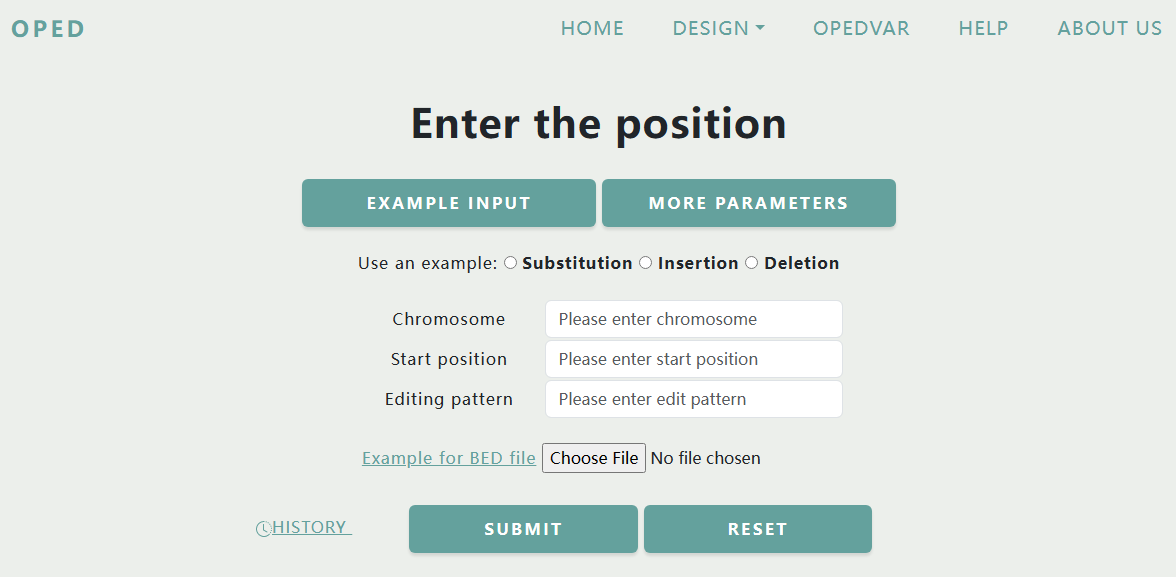
Users need to enter the Chromosome, the Start position of the edited base in the human GRCh38 genome map, and the Editing pattern, or you can submit a BED format file containing these information. You can view the rules for the position by clicking button "Example Input", and check the parameters by "More Parameters".
Similarly to Design by Sequence, we provide users with quick examples. After clicking the radio button of substitution, insertion, or deletion, the corresponding example information of Chromosome, Start position and Editing pattern will be autofilled into the text input boxes.
For the query of the Position, several rules should be followed:
- Chromosome should be written like "chr1" or "chrX".
- Start position refers to the actual location of the edited base in the sequence of a chromosome, it should be a positive integer.
- Editing pattern are similar to rules in sequence. Backslash "/" is used for Substitutions, the plus sign "+" is used for Insertions, and the minus sign "-" is used for Deletions.
For example:
| Type | Chromosome | Start position | Editing pattern |
|---|---|---|---|
| Substitution | chr1 | 943995 | /T |
| Insertion | chr1 | 943995 | +GTATT |
| Deletion | chr1 | 943995 | -GAGAACTCGG |
For BED files, the content needs to meet the following rule:
- BED file should be organized in five columns: Chromosome, StartLocation, EndLocation, EditingPattern and PositionName.
- Columns should be separated by '\t'
- We will take the EndLocation into account and ignore the StartLocation, as the EndLocation represents the actual location of the editing site in the sequence.
We provide an example BED file that can be downloaded by clicking on the "Example for BED file" link.
2.3 Search in OPEDVar
Users can access the OPEDVar through two ways:
a) Navigation Bar -> OPEDVar;
b) HOME Page -> "GO TO OPEDVAR!" Button.
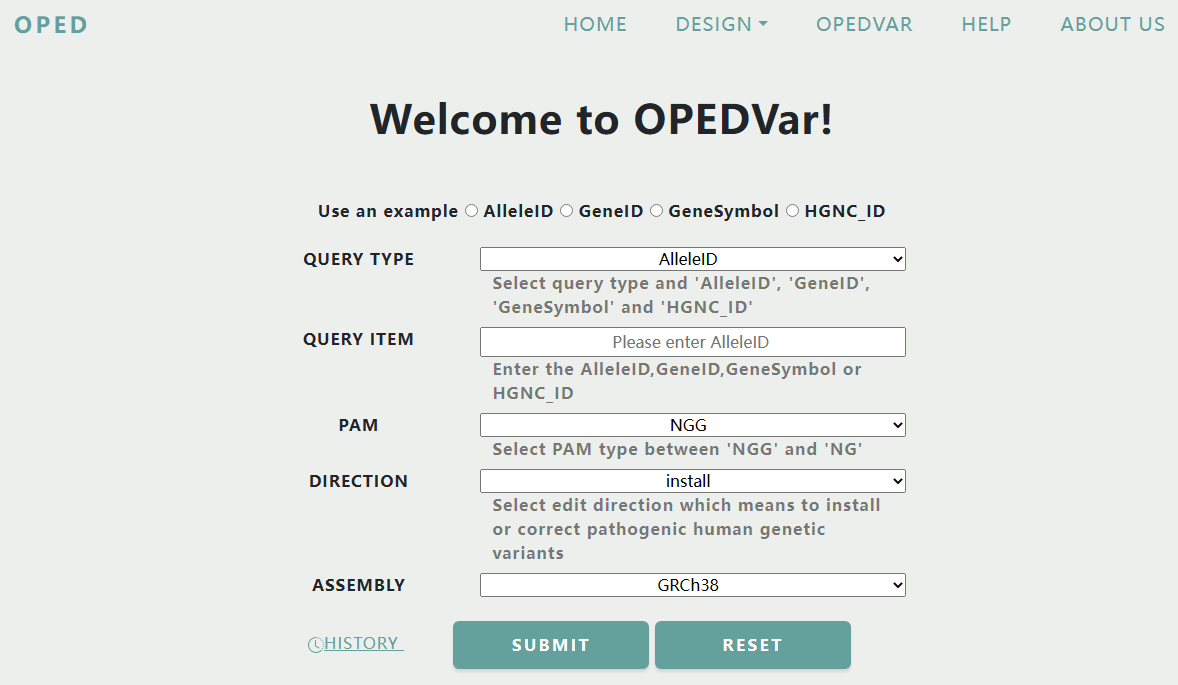
Users can search by AlleleID, GeneID, GeneSymbol and HGNC_ID to obtain optimized pegRNAs and sgRNAs to correct or install pathogenic human genetic variants.
3. Result display
After users submit their queries, OPED will run calculations immediately, and the designed results will be displayed in the form of a table.
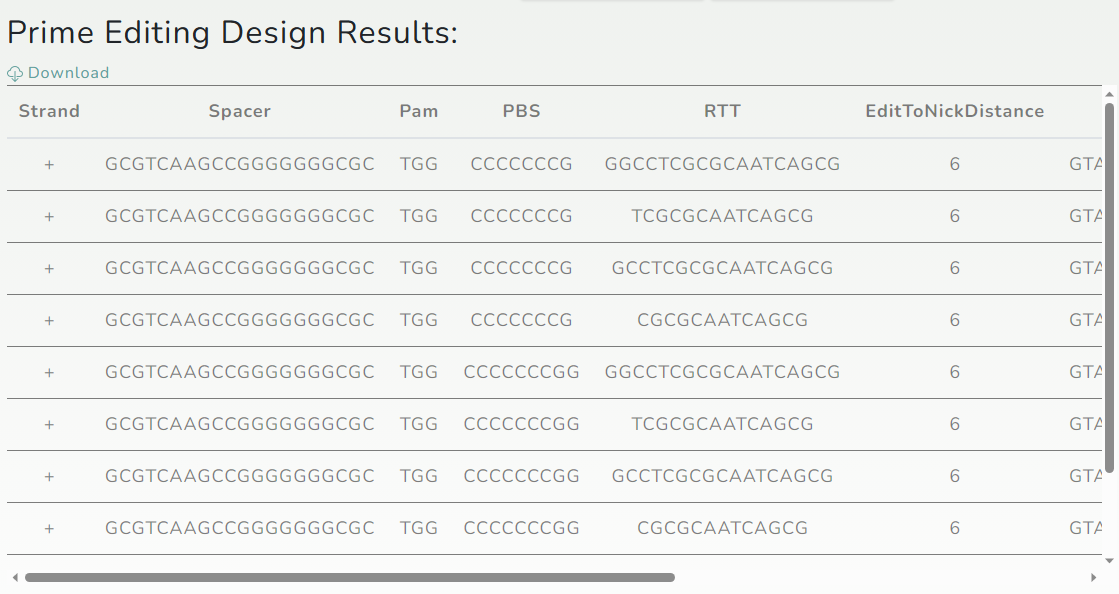
We provide a download function for users, you can download the results table by clicking the
"Download" button at the upper left corner of the table.
At the same time, we provide a function to accessing history records. By clicking the
"HISTORY" link, users can get all the submitted query records, and their detail
information, including sequence information (for sequence queries), position information (for
position queries), gene ID information (for OPEDVar search queries), and corresponding design
parameters they used. Then users get the design results of these queries by clicking the
hyperlink of the records.